| Workpackage 2: Instrumentation Development |

|

|

|
|
Workpackage 2 is the central workpackage of the COMPUTIS project to develop necessary instrumental components. It includes components needed for all of the following workpackages 3 to 6. Key to mass spectrometric imaging is the development of enhanced instrumental techniques for desorption, ionization and detection of biomolecules. The specific objectives of WP2 are:
Optimization of new cluster sourcesThis task included the optimization of new cluster sources for SIMS imaging. CNRS has compared, using rat brain tissue sections (defined as a standard biological sample in WP1), the capabilities of liquid metal ion guns fitted with a gold or a bismuth emitter and fullerene cluster ion sources. Primary ion currents, ion source stability, secondary ion yields, disappearance cross section, efficiency, specific data rate and useful lateral resolution have been evaluated using several secondary ions also defined as standards in WP1. The main conclusion is that the bismuth ion source (patented by Ion-Tof GmbH), when choosing Bi3+ or Bi52+ primary ions, is the best cluster ion source. The capabilities of a C60 fullerene ion source towards depth profiling directly into a rat brain tissue section are topic of D3.3. It has been investigated by FOM whether the improvement in sensitivity provided by a cluster source on native tissue sections is also generated after the tissue sections have been chemically modified by a thin gold coating or matrix deposition. These chemical modification techniques have been independently reported to offer increased molecular ion yields. It was found that the molecular ion yield improvements delivered by surface modification and cluster sources were somewhat additive, and that the most intense signals were obtained by exploiting their combination. Initial attempts to exploit this combination suffered from an unstable cluster source: however, such instability has been addressed by the manufacture and long experiments (high resolution experiments of large areas, of uncoated tissue) have been performed by the CNRS group. 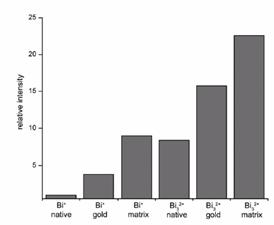 Comparison of ion yield enhancements of the higher mass molecular ions obtained with Bi32+ polyatomic primary ions (with respect to Bi1+ atomic primary ions) for uncoated, gold coated and matrix coated tissue sections.
Microscope detector evaluationThis task included the evaluation of a microscope mode imaging detector that can record (x, y, t) information with 1 ns time resolution and 80 mm spatial resolution, for the purposes of rapid protein imaging. FOM has assembled the delay-line detector and mounted it on a non-imaging MALDI-ToF mass spectrometer to test the data acquisition, control (and optimisation) and processing facilities of the existing software. To test the (x, y, t) capabilities of this detector (on an instrument without the necessary imaging capabilities), a mask was placed in front of the detector assembly, which contains a series of lines of decreasing dimensions. The mask imparts this lined-structure onto the detected ions, thus allowing the imaging capabilities of the (x, y, t) at different resolutions and with different analyte ions. Using a range of ions, including the large protein ions bovine serum albumin and trypsinogen, the (x, y, t) detector has spatially and temporally resolved the lined structures of dimensions > 500 mm with 1 ns time resolution.
One of the major difficulties found during this project, is the limited multi-hit capabilities of the commercial software. Though the detector is designed to allow multi hit capabilities when operated in ADC (Analog-to-Digital-Converter) mode, close examination of the commercial software revealed that it uses very basic pick picking algorithms. This has resulted in the software rejecting the majority of the protein ion signals, and thus limiting the contrast of the images. Two solutions to this problem will be developed in parallel:
Gold implantation and surface coverageThis task included the development of protocols for optimized gold implantation and gold surface coverage for biomolecular imaging. The effect of gold implantation and surface coating on sensitivity; spatial resolution and surface charge build-up was investigated. From these results, a standard protocol was devised to provide optimal sensitivity and spatial resolution for Molecular Imaging Mass Spectrometry (MIMS) of peptides, proteins and other biomolecules. The so-called MILDI method (Matrix Implanted Laser Desorption Ionization) was evaluated. On the one hand, the obtained results were rather discouraging and showed that the sensitivity of the method is not sufficiently high to be further investigated for imaging mass spectrometry. Gold implantation and gold coating were investigated in their ability to improve the sensitivity of SIMS, MALDI and LDI for the analysis of biological samples. The results of FOM demonstrated that gold implantation did not provide sufficient sensitivity for functional imaging mass spectrometry investigations, too. On the other hand, gold surface coverage was extremely promising: it provides significantly more sensitivity, is applicable to both SIMS and MALDI and is compatible with high resolution imaging. In SIMS, the sensitivity enhancement provided by gold coating on a sample is comparable to that obtained by using a cluster source. As indicated in WP2/Task1, the combination of cluster source and gold coating provides the highest yields. These yields were found to be sufficient to investigate the sub-cellular distributions of lipids using their molecular ions. This latter aspect is of vital importance as it enables the analyst to distinguish between lipids and thus investigate the spatial distributions of lipids with more specificity.
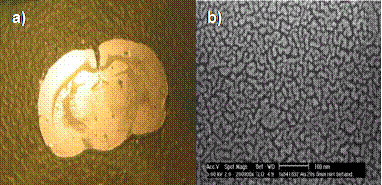 A photograph (a) and SEM micrograph (b) of a rat brain tissue section coated with 1 nm of gold. Scale bar = 100 nm. Under ambient light conditions the gold-coated samples often have a greenish hue
The only drawback of gold coating on a sample is that, with SIMS, it can be difficult to determine which analytes will be detected with higher sensitivity. However, as each experiment can be performed in a few minutes, a simple trial experiment can quickly establish those questions that can benefit from gold coating. The conductivity provided by the gold coating improves the performance of mass analyzers that are sensitive to surface charge, such as the stigmatic ion optical microscope or the time-gate of a TOF/TOF analyzer, to such an extent that it is becoming one of the standard sample preparation treatments in biological imaging mass spectrometry. 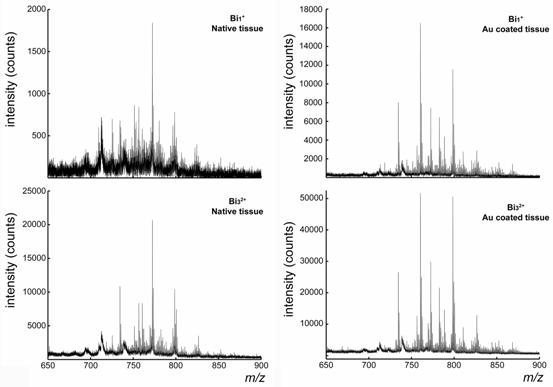 Comparison of the positive-ion SIMS spectra obtained from uncoated and gold-coated tissue section using Bi1+ and Bi32+ primary ions. The spectra concentrate on the lipid region of the mass spectra, and clearly show substantial changes.
Desorption Electrospray Ionisation for MIMSVarious versions of Desorption Ionisation Electrospray (DESI) interfaces were developed and tested. A nitrogen gas assisted version of the imaging DESI source for the COMPUTIS project gave excellent results in comparison to literature (non-imaging) values. It was found however, that the DESI method, even under optimal conditions, cannot be regarded as an advantageous method for imaging purposes as aimed at within the COMPUTIS project.
The limit of detection (LOD) was found to be still much too high under optimized conditions and is not sufficient for imaging real biological samples like tissues and single cells. The usable lateral resolution was found to be rather poor, due to sample migration effects and divergence of the primary ion / spray / gas beam. A minimal usable (effective) lateral resolution of about 1000 µm was found (going down to about 100 to 200 µm with unfavourable deterioration of sensitivity). Substance classes were limited to lipids, small molecules and small peptides. For larger molecules such as proteins, LOD values significantly increased. Biological samples were found to be severely destroyed by the intense nitrogen gas stream under optimized sensitivity conditions. The method thus appears to be unfavourable for sensitive biological samples. In total, the investigation of the DESI method for MS imaging purposes has shown that it is not advisable to further follow the instrumental and methodological developments of DESI imaging within the COMPUTIS project. 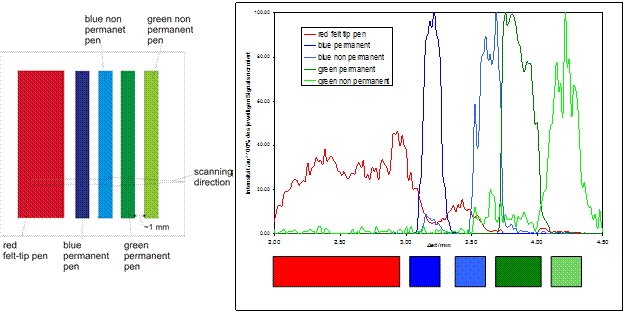 DESI imaging experiment of different felt tip pen entries on PMMA surface. Plate size was 2 cm x 2 cm.
UV-Confocal Microscopy combined with MIMSA new setup of combining scanning Matrix Assisted Laser Desorption Inization (MALDI) with confocal microscopy in the UV range was developed and tested. This setup is shown in the following: 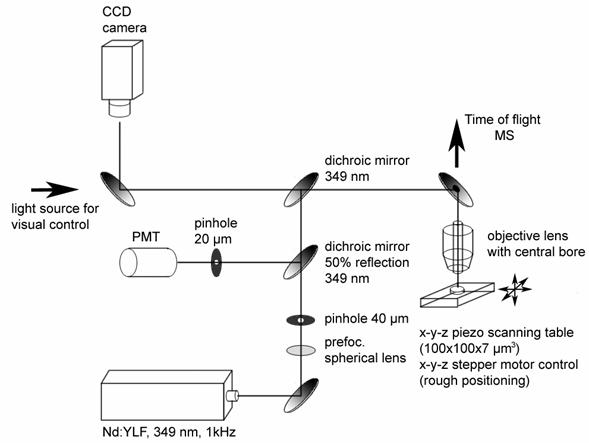 Scheme of the setup for combined confocal MALDI-TOF mass spectrometric imaging and confocal fluorescent imaging Results obtained with the combined setup are demonstrated in the following: 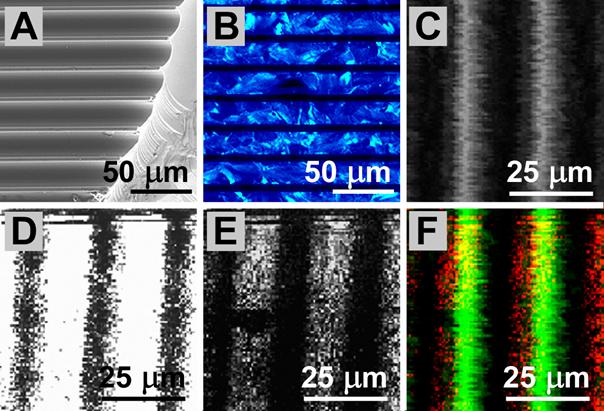 SMALDI and confocal microscopy (CLSM) performance tests. A patterned silicon wafer was used as a test specimen. A: Silicon wafer with line pattern imaged in a conventional scanning electron microscope (SEM). The specimen plane is tilted by 60 degrees against the beam line. The fracture shows the line profile being elliptic. B: The same wafer pattern filled with DHB aqueous solution and air dried to crystallization. The bars are not covered with DHB and thus remain dark. Imaging in a conventional microscope (Olympus BX-40) working in epifluorescence mode. Excitation at 320-380 nm, fluorescence detection 410 nm long pass filter. C: CLSM image obtained from line pattern wafer in the MALDI confocal UV microscope in reflection mode at 349 nm. Illumination with a frequency-tripled Nd:YLF laser. Detection of the reflected light after filtering by a non wavelength-selective beam splitter. The bright lines represent the wafer bars. The dark regions correspond to the grooves etched into the wafer surface. D: SMALDI image of DHB at m/z = 137 u. The same pattern as in C filled with DHB and allowed to dry. The bars are dark while the DHB gives a bright signal. E: As in D but the wafer filled with a mix of DHB as MALDI matrix and the peptide melittin for detection. SMALDI image was composed from signal of melittin at m/z = 2846.8 u ( [M+H]+mono). F: Superposition of C and E. The green channel contains the reflection light signal while the red channel represents the melittin mass spectrometer signal.
Instrumentation for combined imaging and structural analysis of biomoleculesThis task addresses the development of new technical capabilities, concerning the ion generation, ion fragmentation and ion detection, and of optimized preparation protocols, that ensure molecular imaging mass spectrometric (MIMS) measurements of precursor (MS) and fragment ions (MSn).
Three types of combinations of ion sources and mass analyzers were developed, MALDI-TOF, MALDI-(LTQ)-FT-ICR and SIMS-TOF. The MALDI-TOF imaging mass spectrometer at FOM is operating in the microscope mode employing a delay line detector. The imaging MALDI LTQ FTICR combination at JLU operates under atmospheric pressure in the ion source and allows for MSn with highest mass accuracy and resolution. PSD like MS/MS methodologies have been worked on and completed for the third mass spectrometer type, a SIMS-TOF (ToF-SIMS IV, Ion Tof, Münster, Germany) at CNRS. This method could already be applied to biological samples like rat brain sections and showed its potential for structural characterisation of the observed compounds. Using SIMS-TOF for imaging, lateral resolutions on the submicrometer scale can easily be achieved at present combined with MS/MS capabilities. 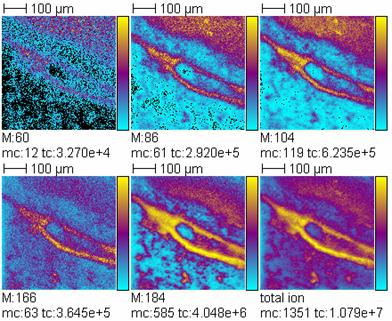 TOF-SIMS images of the precursor ion (phosphocholine headgroup, m/z 184) and its fragment ions (at m/z 60, 86, 104, 166) at the edge of the corpus callosum of a rat brain section deposited on a stainless steel plate without treatment. Field of view 500 x 500 Ám▓; pixel size 1 Ám; compression to 4 Ám; Bi3+ projectiles; primary ion dose density: 1x1012 ions.cm-▓. mc corresponds to the maximal counts on one pixel and tc to the total counts of the image.
Position calibration targetLocalization of specific microareas of a sample, which is under investigation in different instruments (SIMS, MS-SIMS, MALDI microprobe imaging, MALDI microscope imaging) and different laboratories (France, the Netherlands, Germany), is not a simple task. These instruments differ considerably in terms of spatial resolution, field-of-vision and GUI. They are also very different in technical concept and features regarding the development of a common position calibration target. Generic calibration targets with position markers usable by various instruments had to be developed. Several strategies were tested as common position calibration systems. Additional structures had to be added to the standard targets in order to allow for calibration with high resolution. Various structuring technologies were evaluated with respect to requested lateral resolution and accuracy, compatibility with target specifications, manufacturing times, availability, and cost. From the set of available techniques, three main candidates were evaluated in this task, laser etching, photolithography and soft lithography. Due to significant advantages of the soft lithography/micro contact printing technique and due to the limitations of the other techniques, micro contact printing was used as the standard fabrication method for the high resolution position calibration system (the micro-structured area in one corner of the targets). An example of a standard calibration target with additional microstructures for high resolution position calibration is shown in the following.
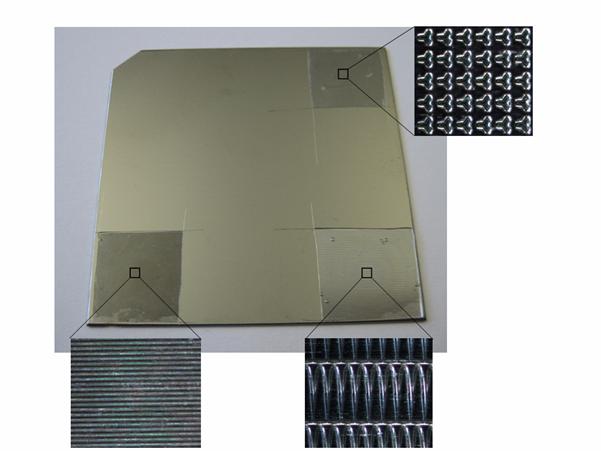 Standard target covered in the corners with cuts of prepared PDMS structures formed by soft lithography Different versions of the patterned concave structures were used, in order to allow their use as optimal position (and orientation and size) calibration markers, as optimal sensitivity calibration structures (lines to be filled with differently concentrated solutions) or as optimal resolution calibration structures (concave saw tooth structures to be filled with analyte solution and to be scanned by MIMS).
|
| < Prev | Next > |
|---|



